
Nanoimager Super-Resolution Microscope
Biological MicroscopyConfocal MicroscopyFluorescent MicroscopyLive Cell ImagingSuper Resolution Microscopy
Understanding the dynamics, trajectories and interactions of single molecules and vesicles in cells presents a fundamental challenge in cell biology. Single-particle tracking (SPT) is a powerful microscopy technique that allows the motion of individual fluorescently-labeled particles (molecules, vesicles, virions or other molecular complexes) to be followed within a medium, or in living cells to investigate behavior.
SPT provides valuable data concerning the positions, paths and interactions of molecules that are highly dynamic or require imaging over extended periods of time. Using sophisticated image processing algorithms, trails of motile particles can be determined with nanometer-precision, and trajectories resolved with millisecond time resolution. This enables track length and diffusion coefficient data to be analyzed quantitatively, applicable for a wide variety of experimental investigations. Moreover, SPT also enables powerful analysis of particle concentration and size distribution for quality control.
To clearly track a protein that has a low abundance in a sample, simply labelling it with a bright and photostable fluorophore is sufficient. However, in order to be able to meet the single-molecule criteria for a protein that is highly expressed, either the concentration of the fluorescent ligand (if you use a tag) needs to be adjusted accordingly, or it is possible to use photoactivatable/convertible proteins, and in turn modulate the intensity of the activation laser to ensure only a subset of molecules are activated simultaneously. This is known as SPT-PALM (Photo-Activated Localisation Microscopy). SPT-PALM is applicable in the context of protein tracking, and even more powerful in the context of EV (extracellular vesicles), which are below 100nm in size.
Extracellular vesicles (EVs) are membrane-bound vesicles secreted by all cells that facilitate intercellular communication. EVs are enriched in surface proteins (such as ALIX, flotillin, and tetraspanins) and contain luminal content (including DNA, RNA, and SiRNA). Importantly, these small nanosized vesicles that transfer active molecules and genetic materials can be isolated from complex biofluids such as blood, urine, and saliva, which makes them ideal candidates for disease diagnostics and therapeutic applications.
Since EVs resemble the composition of their parental origin, secreted vesicles are becoming a promising source for biomarker discovery, as the specific types of surface proteins and cargo may have profound pathophysiological significance. The ability to identify and measure the presence or abundance of particular components could represent particular “disease signatures”. Therefore, characterising molecular profiles of EVs may yield powerful diagnostic capabilities that more accurately determine the onset of distinct cancers1, neurodegeneration2, and cardiovascular diseases3. In this application note, we will discuss how super-resolution microscopy can be used as a powerful tool for EV characterisation and imaging.
Current methods of EV characterisation include flow cytometry, nanoparticle tracking analysis (NTA), epifluorescence imaging techniques and electron microscopy (EM). However, despite the commonplace of these techniques, they often fail to give the necessary resolution to accurately characterise individual EVs or, in the case of EM, struggle to visualise multiple markers in a single-experiment, limiting the ability to assess the molecular signatures of EVs.
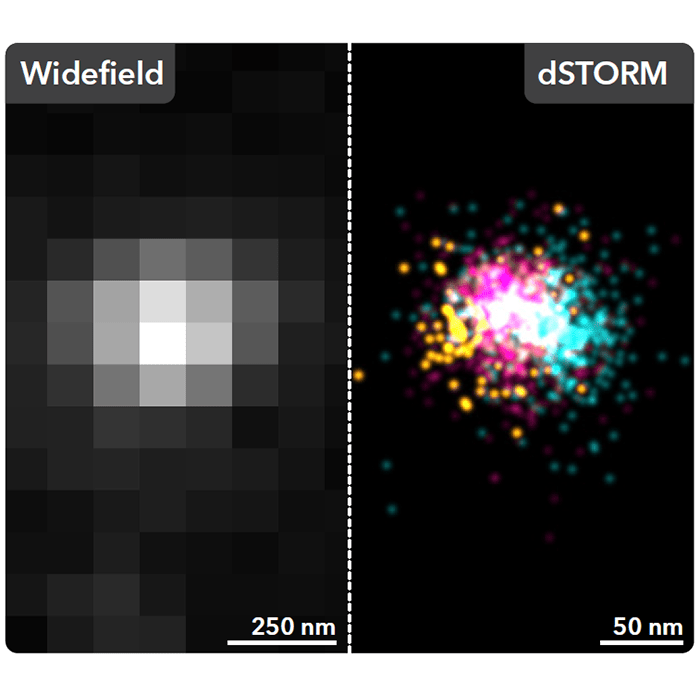
Super-resolution imaging techniques, featured in the MISEV18 guidelines, can overcome the resolution limits associated with conventional light microscopy. Through Single-molecule localisation microscopy (SMLM) techniques, such as dSTORM and PALM, EV biomarkers are labeled with with a class of fluorophores of which the photochemical properties can be exploited to stochastically switch them between a dark and an emissive state. This allows small subsets of fluorophores to be detected in isolation and the localisation of each fluorescent molecule to be fitted with a Gaussian function. By imaging in this way, SMLM retains the advantage associated with traditional fluorescence imaging techniques while circumventing the diffraction-limit, providing an achievable resolution exceeding 20 nm and allowing the spatial distribution of EV markers to be visualised with single-molecule sensitivity.
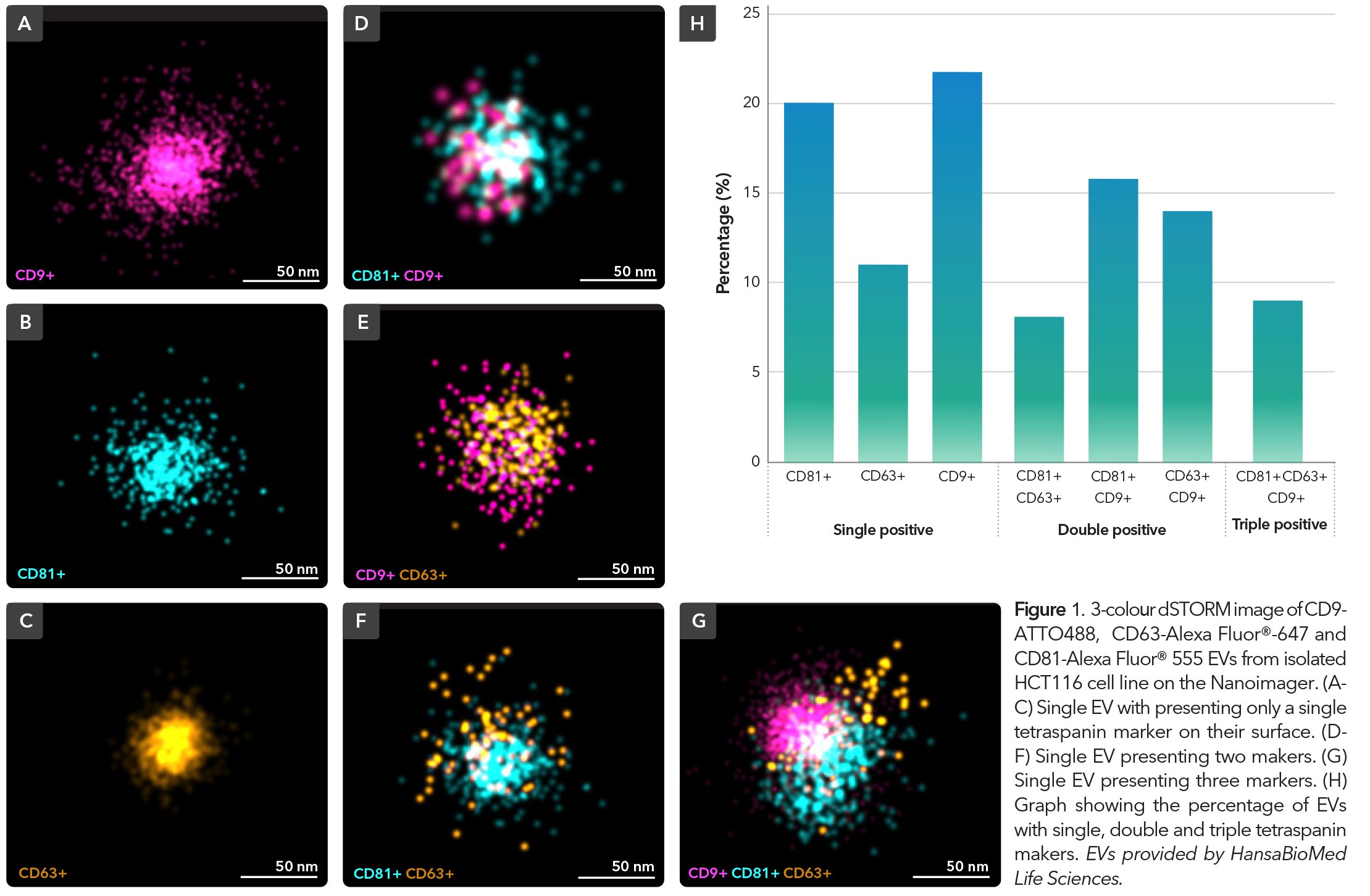
The Nanoimager is a comprehensive SMLM platform for precise and detailed molecular characterisation of EVs. With a multitude of imaging modalities available, this data-driven platform facilitates exploration into composition and behavior of EVs, to provide deeper insights into vesicle subpopulation signaling pathways and intracellular interactions from both a fixed sample and live-cell perspective. To demonstrate the capabilities of SMLM on the Nanoimager in EV characterisation, HCT116 EVs were isolated, purified and immunostained with commercially available antibodies against surface membrane tetraspanins, CD9, CD63 and CD81and imaged using dSTORM (Figure 1). The results demonstrated a heterogeneous population of EVs that were either single (A-C), double (D-F) or triple (G) positive for tetraspanins. Subsequent analysis would allow for the size profiles and relative abundance of each of the markers to be quantified across the EV population.
In addition to the dSTORM profiling, the SPT capability of the Nanoimager allows for tracking dynamics of EVs in order to understand the intracellular transport pathways involved with EV uptake and trafficking.
Here’s an example using human umbilical vein endothelial cells (Figure 2), it’s possible to track the EVs that are stained with NHS-AF488 and image over time to see where they’re diffusing within the cell and whether they’re targeting the nucleus. The dynamic behavior between the EVs close to the nucleus versus the periphery are clearly differentiated by their diffusion coefficients, which represent how mobile they are. Those near the nucleus are less mobile and are more likely interfacing with the nuclear membrane as opposed to those in the periphery. These insights to EV dynamics are not yet possible on other systems. However, with the Nanoimager’s live cell imaging feature along with the SPT feature, researchers can track localisations of EV therapeutic cargos and quantify displacement, directionality, and diffusion speed as they are internalized into cellular compartments. This data highlights the multi-factor characterisation capabilities that follow SMLM imaging, enabling researchers to assess unique protein signatures or changes in biomarker number across populations, which could have important connotations for early disease diagnostics and better understanding EV functionality.
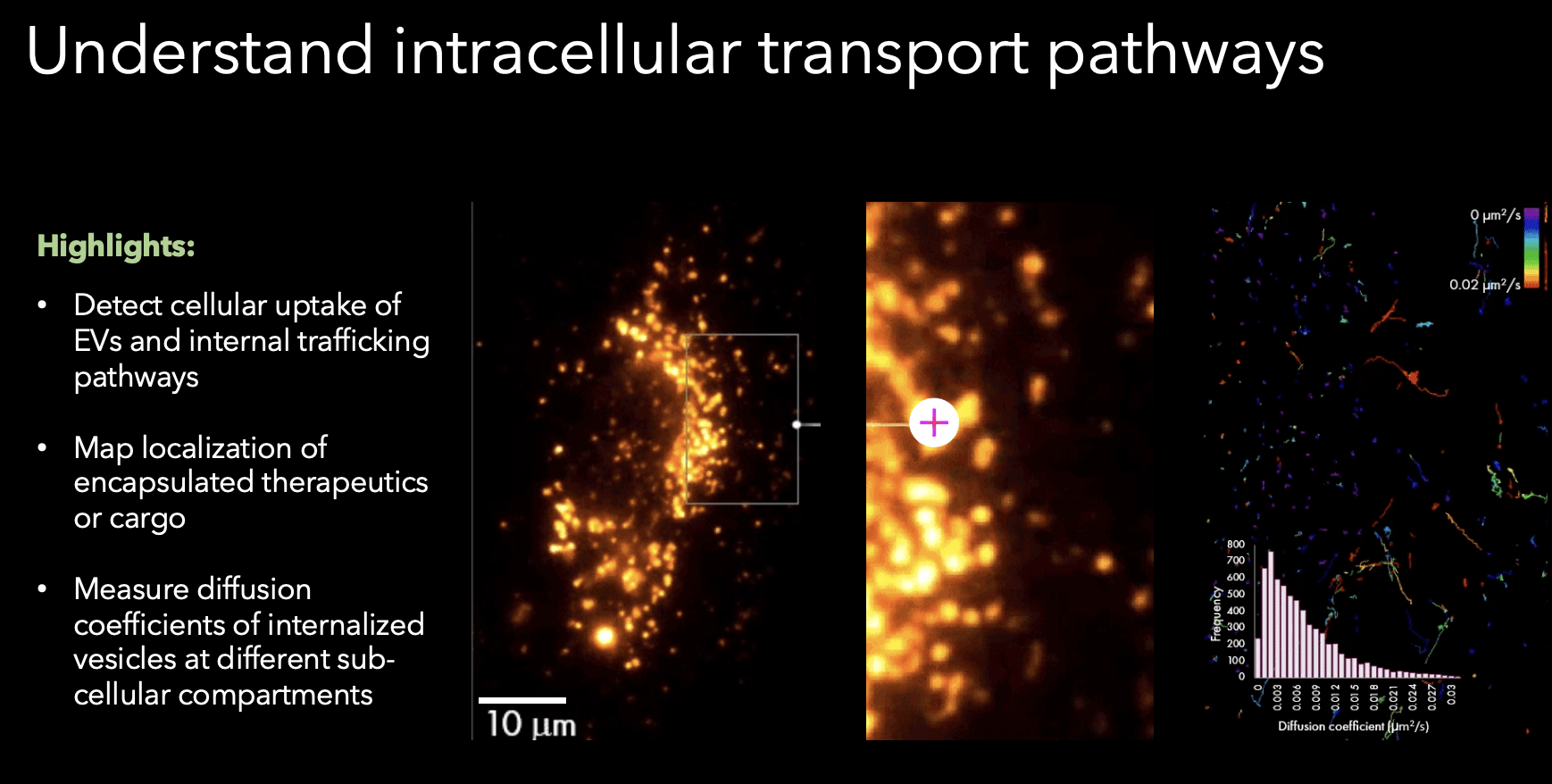
Figure 2. Tracking of EVs NHS-AF488 within a human umbilical vein endothelial cell. Sample from Prof. Christopher Gregory, University of Edinburgh.
